Tissue Study
CS348b final project - Anthony Sherbondy
Goal
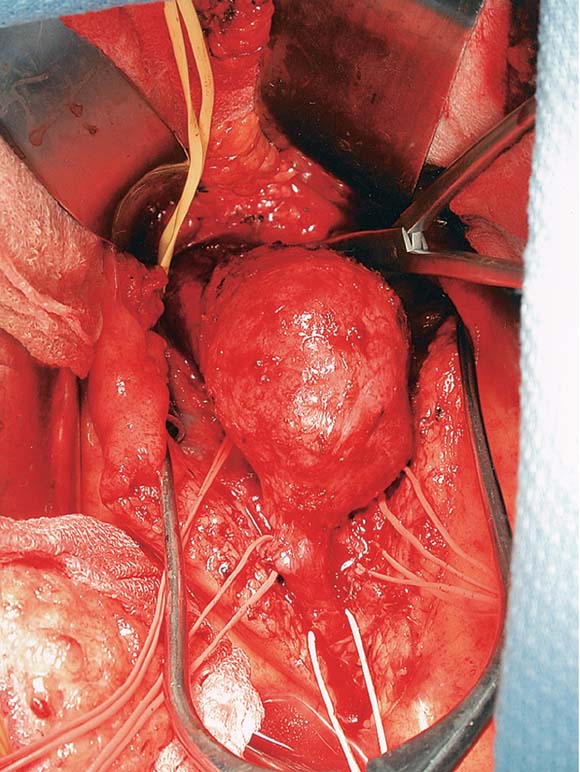
To realistically render scenes involving internal human organs. Specifically, I decided to render a scene based on a picture during an open aortic aneurysm repair. Shown below, courtesy of the Journal of Vascular Surgery.Materials
The data for the anatomy was originally procured from a mult-detector
spiral CT data set of a patient with an abdominal aortic aneurysm.
The data set was anisotropic with ~0.6mm voxel size (in-plane) and
~0.8mm voxel length (through-plane), where through plane describes the
axis that extends from the head to the toes. The images were taking
during contrast enhancement of the blood, thereby giving the aorta a
higher density value then the surrounding soft tissue. An
image of the data set is shown below.Scene Construction
The segmentation of the CT volume image into a scene composed of the interesting surfaces for a likeness to the above surgical picture proved to be more challenging than initially anticipated. Segmentation of the image and scene construction was as follows:- Segment the structure of interest (aorta or supporting tissue) using a previously developed interactive seeded region growing technique as described in the Sherbondy03 paper. The segmentation algorithm was chosen because it smoothly segmented between segmentation regions as the boundary of the segmentation is smoothed with a diffusion metric.
- The aorta segmentation and the background tissue segmentation were saved as seperate images.
- Marching cubes was applied to each segmented image to obtain a triangle mesh representation of the surface.
- Dividing cubes was also run on the segmentation in order to
produce an evenly distributed set of points with the appropriate
distance of one mean free path between sample points
Rendering Algorithm
In order to render the tissue realistically, I decided to use the BSSRDF approximation to light transport. Specifically, I implemented the technique discussed in the Jensen02 paper. The following is a breakdown of the rendering algorithm:- Load the uniformly distributed points and normals of the mesh.
- Estimate irradiance arriving at each sample mesh point by tracing shadow rays.
- Store each mesh point in an octree as described in Jensen02.
- Raytrace the scene.
- For each intersection with the tissue material estimate the
radiance due to multiple scattering as described in Jensen02 and due to
single scattering as described in Jensen01.
Results
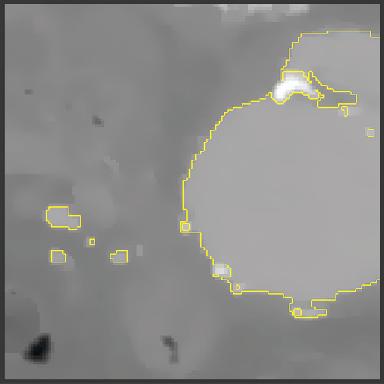
Subsection of CT Image of the aorta and surrounding tissue. The aorta is highlighted with the yellow boundary.
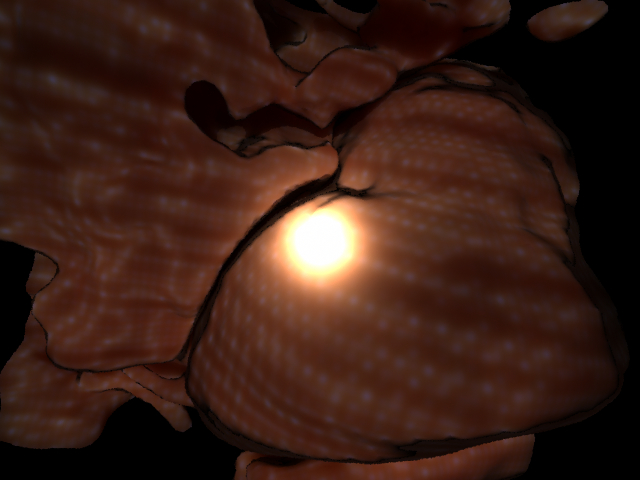
The aorta and some surrounding tissue segmented with the segmentation approach described before. I think the surrounding tissue structure does look as though it might have been pulled around the aorta. I also chose to use a wide field of view lens for this scene as that is what is common in medical practice to show alot of what is happening in the scene at once.
Here is a rendering of the aorta alone after I found the right mesh size of the points. Notice, the banding along the length of the aorta. This is due to the anisotropically sampled CT data. The voxels were sampled (0.5x0.5x0.8mm).
Here is a rendering of the aorta alone after I tried to get the tissue looking more pink as it does inside the body.
Here was supposed to be the final scene looking pretty tissue like, but the image is suffering from the aliasing artifacts due to the diffusion estimation on the poorly sampled point mesh as I describe in the discussion.
Discussion
As previously mentioned the scene construction phase was more difficult then initially anticipated. The segmentation process was challenging not because of segmenting the aorta as that was contrast enhanced and so easily picked out amongst the other tissue, but the segmentation of the background tissue that would be present in a surgical situation is much trickier. As you can see from the CT section figure, the aorta is surrounded by soft-tissue (fat, other vessels, etc.). In an open surgery as shown above, the surgeon would physically displace this tissue on his/her way to the aorta. The segmentation task involved creating a realistic impression of what the tissue would look like if pulled away. This became a somewhat challenging artistic task (for I have little artistic talent) to represent tissue boundaries that were not actually present at the time of the CT scan. I chose to still segment the data to find this imaginary boundary, but to use a segmentation algorithm that would represent a relatively smooth and tissue like boundary appearance when it does not actually have an edge to correspond with the boundary of a tissue. In other words, the region growing methodology with a diffusion based merging metric as described in the Sherbondy03 paper would allow me to introduce tissue like boundaries in a segmentation that does not actually contain a visible boundary at that point.After the segmentation, my troubles were still not finished with the scene construction. After obtaining a polygon representation of my segmentation surfaces by using marching cubes, I initially attempted to use a smoothed version of the vertices of the polygons as the sample points for the irradiance estimates in the rendering algorithm. I thought I had placed the points at a safe distance between each other (i.e. I was sampling with a frequency above the mean free path length ~1.3), however, I did not realize until very late (day before project due date) that I was still not sampling high enough. I needed to raise the sampling frequency and lower the distance between points to 0.6.
In order to verify this theory, I used dividing cubes to create a dense point cloud of my surface instead of the more sparse polygon representation. The point cloud sampled images were generated on the smaller "aorta only" where I generated 250k points. This worked much better as you can see the images of the aorta only are free of the dotted sub-sampling pattern shown in the full scene image.
By solving my sampling problem, I had also inadvertently created a registration problem. My point cloud needed to match the normals of my polygon representation. I could not just get the gradients from the raw CT volume and use those as normals because the normals would not match the ones calculated for the polygons. Instead, I had to write another quick algorithm to match my pointcloud to my surface polygons and create normals from the surface polygon normals to be stored at each point in the point cloud. I only used a nearest neighbor search. But you could imagine that trying to write this algorithm the day before the project due date, I did not have time to apply it to the scene with the background tissue as well. (The main reason is that I couldn't debug a file access issue that had come up that was due to the 700k points I was working with.
As already reported, it was only after debugging the rendering algorithm for awhile, that I realized one of the deficiencies of rendering my scene was due to the poor sampling rate initially chosen. This also made the debugging of the multiple scatter particularly challenging because I was unaware that the model was making the diffusion approximation incorrect. Anyway, the end result is that I was not able to get the scene I wanted appropriately sampled in time.
I did not include my scene construction software. I.e. the segmentation, surface extraction, point extraction and point to surface registration because I implemented that in my software architecture that would require many support files to demonstrate. Needless, to say each core algorithm is documented fully in the literature and I only needed to make particular modifications (although time consuming) to run the algorithms for my situation.
Relavent Works
Henrik Wann Jensen and Juan Buhler. "A Rapid Hierarchical
Rendering Technique for Translucent Materials." In Proceedings of
SIGGRAPH 2002.Henrik Wann Jensen, et. al. "A Practical Model for Subsurface Light Transport." In Proceedings of SIGGRAPH 2003.
Matt Pharr and Pat Hanrahan. "Monte Carlo Evaluation of Non-Linear Scattering Equations for Subsurface Reflection." In Proceedings of SIGGRAPH 2000.
Pat Hanrahan and Wolfgang Krueger. "Reflection from Layered Surfaces due to Subsurface Scattering." Computer Graphics Proceedings. 1993.
Nelson Max. "Optical Models for Direct Volume Rendering." IEEE Transactions on Visualization and Computer Graphics. 1995.
Joe Stam. "Multiple Scattering as a Diffusion Process."
Arnold D. Kim and Joseph B. Keller. "Light Propogation in Biological Tissue." Journal of the Optical Society of America 2003.
Anthony Sherbondy, Mike Houston and Sandy Napel. "Fast Volume Segmentation with Simultaneous Visualization Using Programmable Graphics Hardware." To Appear in IEEE Visualization 2003.